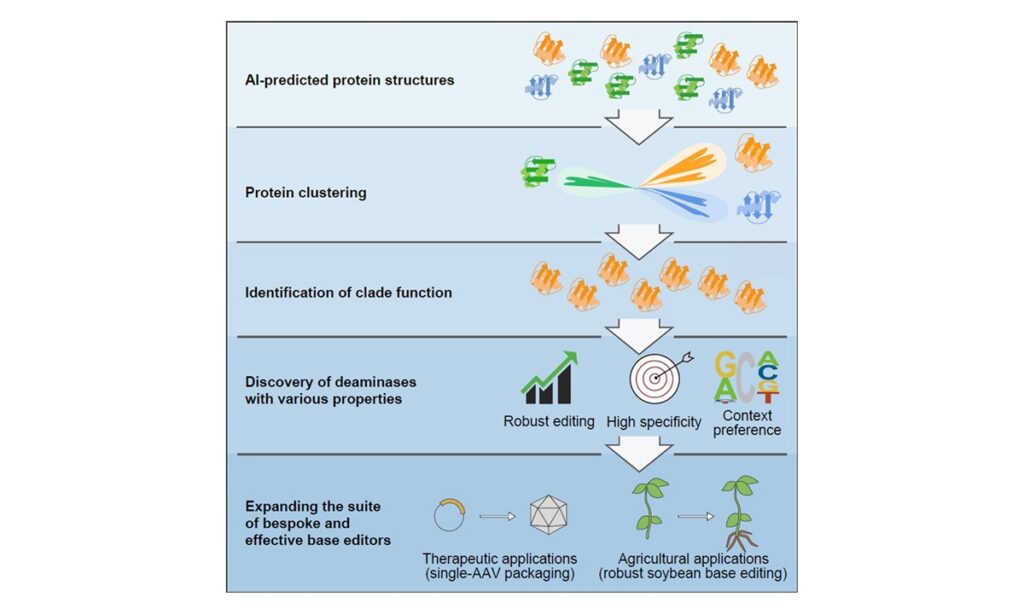
Chinese researchers have developed the use of artificial intelligence (AI)-assisted methods to discover novel deaminase proteins with unique functions through structural prediction and classification, which will help China gain a competitive advantage in the future biotechnology industry, the Institute of Genetics and Developmental Biology (IGDB) of the Chinese Academy of Sciences said on Wednesday.
This approach has “opened up a range of applications for the discovery and creation of desired plant genetic traits,” reads a statement by the IGDB. The results were published on June 27 in Cell magazine.
AI-guided structural classification establishes new deaminase family relationships, Gao Caixia’s research group wrote in the published article. These discovered deaminases, based on AI-assisted structural predictions, expand the utility of base editors for various therapeutic or agricultural breeding efforts in the future greatly, according to the article.
The newly developed base editing system is a precision genome editing tech built on China’s independent intellectual property rights. Researchers have submitted patent application through the Patent Cooperation Treaty, which is expected to break the patent monopoly of base editing, according to the IGDB.
Gao’s group conducts research in three major areas – plant genome editing technology, biological secure new breeding technology, and genome editing directed molecular design breeding, according to the lab’s official website.
The study was supported by the National Natural Science Foundation of China, the National Key Research and Development Program of China and the Ministry of Agriculture and Rural Affairs of China, among others, according to the IGDB.
(Global Times)